Efficiently handle slightly big data with Apache Arrow in R
R Apache Big DataIn systems biology, we often need to work with slightly big data. Not so big to justify setting up a database or using a high-performance cluster, but still a bit too big to comfortably work with in memory. We are talking about files in the 10 to 500 GB range, such as:
- Omics data like RNAseq or proteomics
- Single-cell phenotype data from high-content microscopy
- Large public data repositories, like the Human Cell Atlas
The Arrow package for R lets us keep our data set on disk, dynamically loading only the rows and columns needed for our analysis. We specify a set of instructions with regular dplyr verbs (filter(), group_by(), summarise() etc.). Then, when we call collect(), the full set of instructions is optimised and handled by Arrows’s efficient back end, returning only the final result into R’s memory.
I was recently working with a 20 GB table containing the combined results of many differential expression analyses. I performed the original analysis on a high-performance cluster, but now I just wanted to summarise and visualise the results. Simply loading the file across the university’s network took around 10 minutes and used up most of my computer’s RAM, making my standard-issue Dell desktop even more sluggish than usual.
Let’s demonstrate the capabilities of Arrow with a small toy data set:
data = tibble(condition = rep(LETTERS[1:10], each = 1000),
gene = rep(1:1000, 10), # Some made up gene ids
fold_change = rnorm(10 * 1000),
p_value = runif(10 * 1000))
data
## # A tibble: 10,000 × 4
## condition gene fold_change p_value
## <chr> <int> <dbl> <dbl>
## 1 A 1 -0.993 0.229
## 2 A 2 2.89 0.182
## 3 A 3 0.138 0.115
## 4 A 4 0.0956 0.934
## 5 A 5 -0.313 0.503
## 6 A 6 1.57 0.704
## 7 A 7 1.51 0.561
## 8 A 8 -0.568 0.763
## 9 A 9 -1.90 0.822
## 10 A 10 -1.40 0.738
## # ℹ 9,990 more rows
# Get the size of the table in memory
object.size(data)
## 281800 bytes
First we save our data as a data as a set of parquet files. By grouping our tibble by the variable condition, Arrow will automatically split the our data into 26 parquet files, each of which contains the data for a single condition. This means we can more efficiently load data for individual conditions in our downstream analysis.
library(arrow)
# As a folder of chunked files
data %>%
group_by(condition) %>%
arrow::write_dataset("data")
# Remove the tibble from memory
rm(data)
The folder structure is as follows:
## data
## ├── condition=A
## │ └── part-0.parquet
## ├── condition=B
## │ └── part-0.parquet
## ├── condition=C
## │ └── part-0.parquet
## ├── condition=D
## │ └── part-0.parquet
## ├── condition=E
## │ └── part-0.parquet
## ├── condition=F
## │ └── part-0.parquet
## ├── condition=G
## │ └── part-0.parquet
## ├── condition=H
## │ └── part-0.parquet
## ├── condition=I
## │ └── part-0.parquet
## └── condition=J
## └── part-0.parquet
Next, we load the dataset back into memory with open_dataset(). This doesn’t actually load in the data, but creates a ’lazy’ table object containing information on the format of the data and a link to the filesystem.
data = open_dataset("data")
# Show the format of the object and its size on in memory
print(data)
## FileSystemDataset with 10 Parquet files
## 4 columns
## gene: int32
## fold_change: double
## p_value: double
## condition: string
object.size(data)
## 504 bytes
As you can see, the table now occupies only 504 bytes in RAM, down from 281800. That’s a 99.8% memory saving!
We can now work with the object using dplyr verbs, just as we would with a regular tibble. Let’s say we want to get the percentage of significantly regulated genes per condition and display the result in a bar chart:
data %>%
filter(p_value < 0.05) %>%
group_by(condition) %>%
summarise(significant_genes = n()) %>%
# Up to this point, the commands have been recorded but no processing has yet occurred.
collect() %>%
ggplot() +
aes(significant_genes, condition) +
geom_bar(stat = 'identity') +
theme_linedraw()
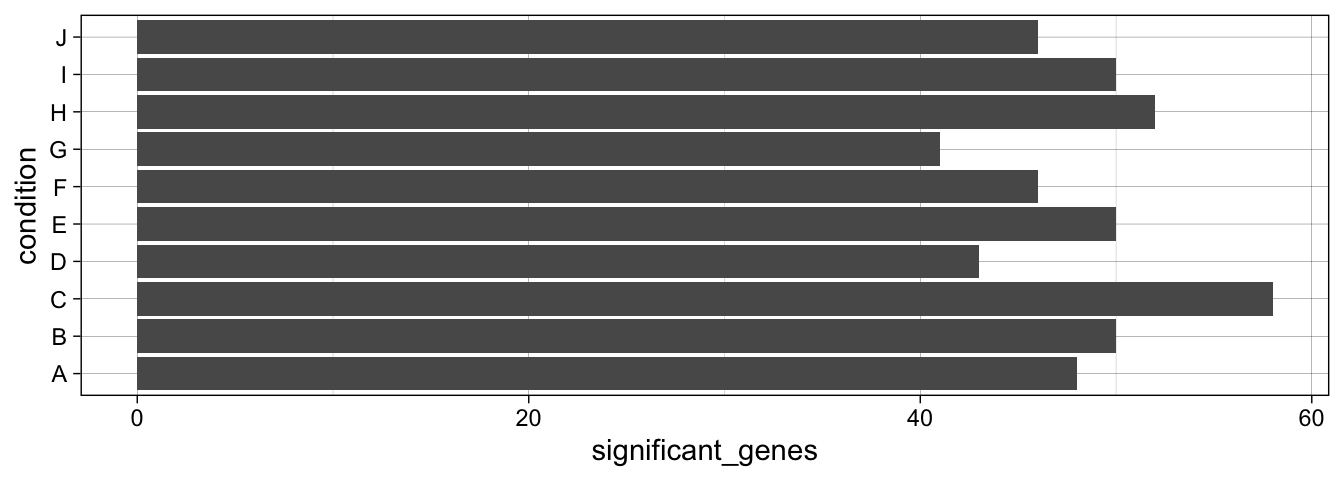
Condition G had the highest number of significant hits. Let’s say we want to take a closer look and get the names of genes significantly up-regulated in condition G. Again, we don’t have to load the whole data set into memory.
data %>%
filter(condition == "G",
p_value < 0.05) %>%
arrange(desc(fold_change)) %>%
head %>%
collect()
## # A tibble: 6 × 4
## gene fold_change p_value condition
## <int> <dbl> <dbl> <chr>
## 1 795 2.18 0.0311 G
## 2 930 1.11 0.0387 G
## 3 418 1.05 0.0426 G
## 4 943 0.989 0.0494 G
## 5 953 0.879 0.0174 G
## 6 583 0.718 0.0348 G
We can directly compare the time it takes to use Arrow vs loading the data from a .csv into memory and running the analysis directly.
library(tictoc)
tic()
result <- read_csv("data.csv") %>%
filter(condition == "G",
p_value < 0.05) %>%
arrange(desc(fold_change)) %>%
head()
toc()
## 0.054 sec elapsed
tic()
result <- open_dataset("data") %>%
filter(condition == "G",
p_value < 0.05) %>%
arrange(desc(fold_change)) %>%
head() %>%
collect()
toc()
## 0.012 sec elapsed
An impressive time saving, especially when scaled up to much larger real-life data sets.
Arrow has many other options and features that I hope to post about in the future, including:
-
Cross-platform compatibility, meaning you can load the same parquet dataset directly into Python
-
Integration with database packages such as
sparklyrandDuckDB, which gives you access to a full range of SQL and optimised machine learning commands
For further reading, check out these other great articles:
-
Manipulate big data with Arrow & DuckDB (Great explanation of how these packages function behind the scenes)
Thanks for reading my first blog post. If you have any feedback, get in touch using the links in the footer.
sessionInfo()
## R version 4.3.3 (2024-02-29)
## Platform: aarch64-apple-darwin20 (64-bit)
## Running under: macOS Sonoma 14.4.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: Europe/Zurich
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] tictoc_1.2.1 arrow_15.0.2.9000 lubridate_1.9.3 forcats_1.0.0
## [5] stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
## [9] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] sass_0.4.9 utf8_1.2.4 generics_0.1.3 blogdown_1.19.1
## [5] stringi_1.8.4 hms_1.1.3 digest_0.6.35 magrittr_2.0.3
## [9] evaluate_0.23 grid_4.3.3 timechange_0.3.0 bookdown_0.39
## [13] fastmap_1.1.1 jsonlite_1.8.8 fansi_1.0.6 scales_1.3.0
## [17] jquerylib_0.1.4 cli_3.6.2 rlang_1.1.3 crayon_1.5.2
## [21] bit64_4.0.5 munsell_0.5.1 withr_3.0.0 cachem_1.0.8
## [25] yaml_2.3.8 parallel_4.3.3 tools_4.3.3 tzdb_0.4.0
## [29] colorspace_2.1-0 assertthat_0.2.1 vctrs_0.6.5 R6_2.5.1
## [33] lifecycle_1.0.4 fs_1.6.4 bit_4.0.5 vroom_1.6.5
## [37] pkgconfig_2.0.3 pillar_1.9.0 bslib_0.7.0 gtable_0.3.5
## [41] glue_1.7.0 highr_0.10 xfun_0.43 tidyselect_1.2.1
## [45] rstudioapi_0.16.0 knitr_1.45 farver_2.1.1 htmltools_0.5.8.1
## [49] labeling_0.4.3 rmarkdown_2.26 compiler_4.3.3